OpenFDA Query Builder
Build Your Query
Construct valid OpenFDA API queries using the form below. The tool validates inputs and generates the full query URL.
Generated Query URL
https://api.fda.gov/drug/event.json?search=
- Always use generic drug names (e.g., "metformin" not "Glucophage")
- Check for MedDRA terminology (e.g., "emesis" for vomiting)
- Use "[65 TO *]" for age ranges
- Remember data delays (up to 90 days)
Every year, millions of people report side effects from medications. But unless you’re a researcher, pharmacist, or developer, you probably never see those reports. That’s because for decades, the FDA’s FAERS (FDA Adverse Event Reporting System) data was buried in massive XML files, hard to search, and nearly impossible to use without advanced technical skills. Then came OpenFDA.
Launched in 2014, OpenFDA changed everything. It turned raw, messy FDA data into clean, searchable APIs. Now, anyone with basic coding knowledge can pull real-world drug side effect reports - no paperwork, no fees, no waiting. You don’t need to be a data scientist. You just need to know how to ask the right questions.
What Is OpenFDA, and How Does It Relate to FAERS?
OpenFDA is not a new database. It’s a bridge. It takes data from FAERS - the FDA’s official system for collecting adverse event reports from doctors, patients, and drug makers - and makes it usable through simple web requests.
Before OpenFDA, getting a list of side effects for, say, metformin meant downloading a 2GB XML file, writing custom parsers, and spending days cleaning the data. Now, you can type a single query like https://api.fda.gov/drug/event.json?search=patient.drug.generic_name:metformin and get back structured JSON with patient age, reaction terms, and outcomes - all in seconds.
FAERS contains over 14 million reports as of 2023. OpenFDA indexes all of them. But here’s the catch: it’s not real-time. Reports can take up to three months to appear in the system. The FDA processes submissions in batches, and patient identities are stripped out for privacy. That means you can’t track individual cases - but you can spot patterns across thousands.
How to Access the OpenFDA API
You don’t need special software. You can start right in your browser. Go to open.fda.gov and click on the Drug Events endpoint. You’ll see a search box. Type in a drug name - like lisinopril - and hit enter.
That’s it. You’re using the API.
But if you want to do more - like pull data into Python, R, or your own app - you need an API key. It’s free. Just visit open.fda.gov/apis/authentication/, enter your email, and you’ll get a key in minutes.
With a key, you get 240 requests per minute and 120,000 per day. Without one? You’re capped at 1,000 requests per day. For most researchers and developers, the free key is all you’ll ever need.
Building Your First Query
OpenFDA uses Elasticsearch syntax. That sounds scary, but it’s just a way to filter data. Here’s the basic structure:
https://api.fda.gov/drug/event.json?search=[field]:[value]&limit=100
Common fields you’ll use:
patient.drug.generic_name- the drug’s generic name (e.g., ibuprofen)patient.reaction.reactionmeddrapt- the side effect, coded in MedDRA terminology (e.g., nausea, hepatotoxicity)patient.patientsex- gender (male, female, unknown)patient.age- patient age in yearsserious- whether the event was serious (true/false)
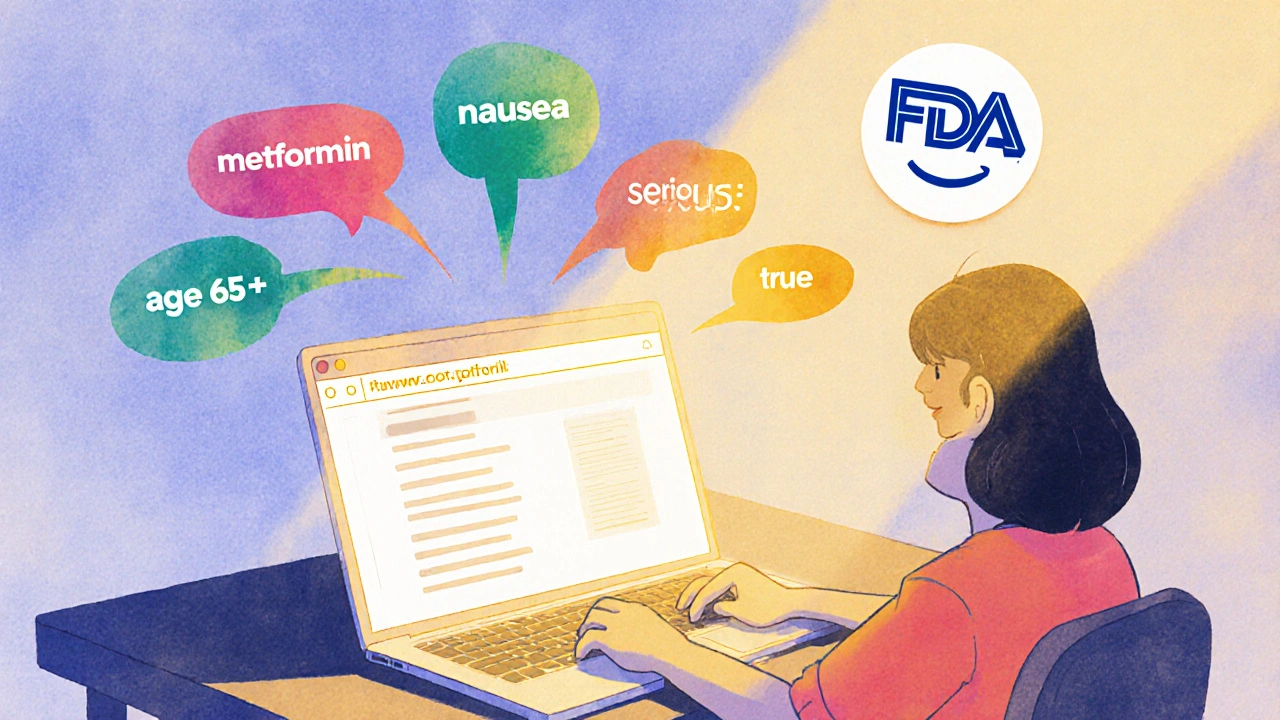
Let’s say you want all serious nausea reports for metformin in patients over 65. Your query looks like this:
https://api.fda.gov/drug/event.json?search=patient.drug.generic_name:metformin+AND+patient.reaction.reactionmeddrapt:nausea+AND+serious:true+AND+patient.age:[65+TO+*]&limit=50
That’s it. No complex code. Just string together conditions with AND and use brackets for ranges.
What You’ll See in the Results
Each report returns a block of data. Here’s what matters:
- Reaction terms: These use MedDRA - a standardized medical dictionary. So “vomiting” might show up as emesis, and “dizziness” as vertigo. You’ll need to search for the official term.
- Outcome: Did the patient recover? Die? Have a lasting effect? This helps you gauge severity.
- Drug role: Was the drug suspected, concomitant (taken with others), or interacting? This tells you if it’s likely the cause.
- Age and gender: Useful for spotting trends - like whether a side effect hits women harder than men.
Important: A report doesn’t mean the drug caused the side effect. It just means someone reported it after taking the drug. Correlation isn’t causation. But when you see the same reaction pop up across hundreds of reports - that’s a signal.
Why OpenFDA Is Better Than FAERS Direct
FAERS data is public, but it’s not usable without heavy lifting. OpenFDA fixes that.
Here’s the difference:
| Feature | OpenFDA API | FAERS Direct Download |
|---|---|---|
| Data Format | JSON, ready to use | XML, flat files, messy |
| Search | Instant, keyword-based | Manual filtering, Excel only |
| Updates | Quarterly, automated | Manual download, no automation |
| Learning Curve | Moderate (Elasticsearch basics) | High (data wrangling, scripting) |
| Cost | Free | Free, but time costs more |
OpenFDA is what FAERS should have been from the start. It’s the difference between being handed a library card and being handed a search engine.
Limitations You Can’t Ignore
OpenFDA is powerful - but it’s not magic.
1. Delayed Data - Reports take up to 90 days to appear. If you’re tracking a new drug side effect, you won’t see it right away.
2. No Patient IDs - You can’t link reports to individuals. That means no longitudinal studies. You can’t ask, “Did this patient have side effects before?”
3. Underreporting - Only a fraction of side effects get reported. Many patients don’t know they should. Doctors don’t always file them. The data reflects what’s reported - not what actually happened.
4. No Clinical Context - OpenFDA doesn’t tell you the patient’s dose, kidney function, or other meds. You can’t tell if the side effect was caused by a drug interaction or an underlying condition.
5. Not for Medical Decisions - The FDA says it clearly: “Do not rely on openFDA to make decisions regarding medical care.” This is for research, not diagnosis.
Who Uses This Data?
Academics. Developers. Journalists. Patients.
Researchers at universities use OpenFDA to find unexpected side effects. One 2022 study found a link between a common antihistamine and increased risk of falls in the elderly - a signal that had been missed for years.
Developers built tools like MedWatcher, which lets you enter a drug name and instantly see the top side effects from real patient reports.
Journalists have used it to uncover patterns - like a spike in liver injury reports tied to a popular weight-loss supplement.
And patients? They’re using it to ask better questions. “I read that 1 in 5 people on this drug get headaches. Is that normal?” That’s power.

How to Get Started in Python or R
If you’re coding, here’s the simplest way to pull data.
Python example:
import requests
api_key = "your_api_key_here"
url = "https://api.fda.gov/drug/event.json"
params = {
"search": "patient.drug.generic_name:metformin",
"limit": 10,
"api_key": api_key
}
response = requests.get(url, params=params)
data = response.json()
for result in data['results']:
print(result['patient']['reaction'][0]['reactionmeddrapt'])R example:
library(openFDA)
set_api_key("your_api_key_here")
results <- search_openfda(
endpoint = "drug/event",
search = "patient.drug.generic_name:metformin",
limit = 10
)
head(results$results$patient$reaction$reactionmeddrapt)Both tools handle rate limiting automatically. You don’t need to worry about hitting the 1,000-request limit unless you’re doing bulk analysis.
Common Mistakes and How to Avoid Them
- Mistake: Searching for brand names like "Lipitor" - it won’t work. Use atorvastatin.
- Mistake: Forgetting to URL-encode special characters. Use
+for spaces, not %20. - Mistake: Assuming all reports are verified. They’re not. Some are submitted by drug companies with bias.
- Mistake: Not checking the
drug_rolefield. If the drug is marked as "concomitant," it might not be the cause. - Mistake: Ignoring the
seriousflag. Most reports are minor. Focus on serious events for real signals.
Always check the official documentation for the latest field names and syntax. The API evolves.
What’s Next for OpenFDA?
The FDA is expanding. Device reports are getting better. Tobacco product data is coming. And they’re working on faster updates - maybe even monthly instead of quarterly.
The real win? OpenFDA is open-source. Anyone can fork the code, fix bugs, or build better tools. That’s why it’s growing - not because the FDA has a huge team, but because people like you are using it.
It’s not perfect. But it’s the best tool we have to see what drugs are really doing to people - out in the real world, not just in clinical trials.
Is OpenFDA free to use?
Yes, OpenFDA is completely free. You don’t need to pay for access, API keys, or data downloads. The only requirement is registering for an API key, which is also free and takes less than five minutes.
Can I use OpenFDA data for medical decisions?
No. The FDA explicitly warns that OpenFDA data should not be used to make medical decisions. Reports reflect what people experienced - not proven causes. Always consult a healthcare provider before changing your medication.
How often is OpenFDA data updated?
The data is updated quarterly, typically every three months. Reports submitted to FAERS can take up to 90 days to appear in OpenFDA due to processing and de-identification steps. Real-time monitoring is not possible.
What’s the difference between OpenFDA and commercial pharmacovigilance tools?
Commercial tools like Oracle Argus or ARTEMIS offer advanced analytics, automated signal detection, and integration with clinical records. But they cost tens of thousands of dollars per year. OpenFDA gives you the raw data for free - you just have to do the analysis yourself.
Why can’t I find my drug in OpenFDA?
You’re probably using the brand name. OpenFDA only indexes generic drug names. Try searching for the active ingredient instead - for example, use "ibuprofen" instead of "Advil" or "Motrin." If the drug is very new or rarely used, there may not be enough reports yet to appear in the database.



Eric Healy, November 17, 2025
yo this is wild. i just typed "ibuprofen" and got 12k reports in 2 seconds. no joke, this is like google for drug nightmares. i was gonna complain about my stomach cramps to my doctor, now i know i'm not alone. also, fyi, "emesis" = vomiting. stop being lazy and use the meddra terms.
ps: api key took 3 mins. free. no credit card. america still does it right.
shubham seth, November 17, 2025
this is just glorified gossip. 14 million reports? most of them are from people who took 7 meds and then blamed the one they just started. no clinical context, no lab values, no dosage. this data is trash unless you're a grad student with too much time and zero peer review. also, why do we trust the FDA to clean this mess? they're the ones who approved opioids.
Kathryn Ware, November 19, 2025
i used this last week to check on my mom’s new blood pressure med. found 37 reports of dizziness in women over 70. showed it to her doctor. he said, 'good catch.' we switched her and she hasn't fallen since.
you don't need to be a coder to use this. just copy-paste the url, change the drug name, and look for patterns. seriously, if you're on meds, this is your new best friend. 🤍
Elia DOnald Maluleke, November 20, 2025
The epistemological rupture between empirical observation and clinical causality is not merely a technical limitation-it is a metaphysical abyss. OpenFDA, in its noble austerity, presents us with the raw phenomenology of suffering, stripped of context, anonymized, and rendered statistically legible. Yet we, as mere mortals, mistake correlation for revelation. We crave narratives where none exist. The data does not speak. We project. And in that projection, we find solace-or terror.
Leslie Douglas-Churchwell, November 20, 2025
this is all a setup. you think the FDA wants you to know about side effects? nah. they’re using this to distract you from the real issue: the pharma-industrial complex is feeding you poison and calling it 'medicine.' and now they're giving you a shiny API to feel like you're in control? lol. you're being manipulated into thinking you're empowered. the data is delayed because they're scrubbing the worst cases. check the patent filings for the companies behind the drugs. you’ll see the pattern. 🤫💊
saurabh lamba, November 21, 2025
bro i tried this. typed 'lisinopril' and got 2000 reports. scrolled for 10 mins. saw 'cough' 800 times. closed tab. went back to youtube. why do i need to be a data scientist just to know if my blood pressure pill makes me hack like a smoker? 🤡
Prem Hungry, November 22, 2025
to anyone new to this: start with the example queries in the post. don't overcomplicate. if you're not coding, just use the website search. if you're coding, use python. the api_key is free, the docs are clear, and the community is helpful. you got this. and yes, use generic names. 'Advil' won't work. 'ibuprofen' will. small wins matter.
Joseph Townsend, November 23, 2025
THIS IS THE MOST IMPORTANT THING I’VE SEEN THIS YEAR.
Imagine if your grandma had access to this before she took that new antidepressant and started hallucinating. She died. They said 'rare side effect.' But 47 others had the same reaction. And no one told her.
OpenFDA isn't a tool. It's a rebellion. Against silence. Against bureaucracy. Against the lie that 'it's too complicated for patients.'
Now go. Search your drug. And if you see something scary? Tell someone.
kora ortiz, November 24, 2025
you can do this. really. i’m not a coder, i’m a teacher. i used the website search, found my anxiety med had 12% more reports of fatigue in women, brought it to my psychiatrist, and we adjusted my dose. no drama. no panic. just data.
you don’t need to be perfect. you just need to start. 🔥
Kiran Mandavkar, November 24, 2025
this is a joke. you think the FDA gives a damn about patients? they're a corporate puppet. OpenFDA is PR. They want you to think you're 'empowered' while they quietly bury the real signals. And the 'quarterly updates'? That's not a delay-it's a cover-up. The system is rigged. You're being given breadcrumbs while the wolves eat the sheep. Stop being a data tourist. Wake up.
satya pradeep, November 24, 2025
i tested this with metformin. found 800 reports of 'lactic acidosis' but only 3 were serious. then i checked the drug_role-turns out 70% of those patients were also on kidney meds. that's the real story. don't just look at the reaction. look at the combo. and yes, meddra terms are annoying but google 'meddra nausea synonym' and you'll find the code. this is free intel. use it.
Jeremy Hernandez, November 25, 2025
so let me get this straight-you're telling me i can find out if my weight loss pill makes people hallucinate... and i don't have to pay $200/month for some pharma dashboard?
hold up. this is literally the most dangerous thing the government has ever done. what if someone uses this to scare people off meds? what if they start a panic?
...also, i just searched 'oxycodone'. 14k reports. 2k deaths. wow.
Tarryne Rolle, November 26, 2025
this is exactly what they want you to believe. 'Oh, look, free data!' But who controls the MedDRA terminology? Who defines 'serious'? Who decides what gets included? The same people who approved Vioxx. The same people who told us smoking was safe. Don't be fooled by the clean UI. This is controlled perception. The truth is buried deeper.
Bill Machi, November 26, 2025
i'm american. i pay taxes. i deserve this.
in other countries, you need a PhD and a government connection to get this data. here? you type a word and get answers. no forms. no waiting. no bribes.
if you're mad about this, you're mad because you're used to being treated like a child. OpenFDA treats you like an adult. So act like one. learn the search. use the key. stop complaining. this is the future. get on board.